Genes, 2020.
Authors: Pillay S, Giandhari J, Tegally H, Wilkinson E, Chimukangara B, Lessells R, Mattison S, Moosa Y, Gazy I, Fish M, Singh L, Khanyile KS, Fonseca V, Giovanetti M, Alcantara LCJ, de Oliveira T.
Journal: Genes,doi: https://doi.org/10.3390/genes11080949:11(8), 949 (2020)
Abstract
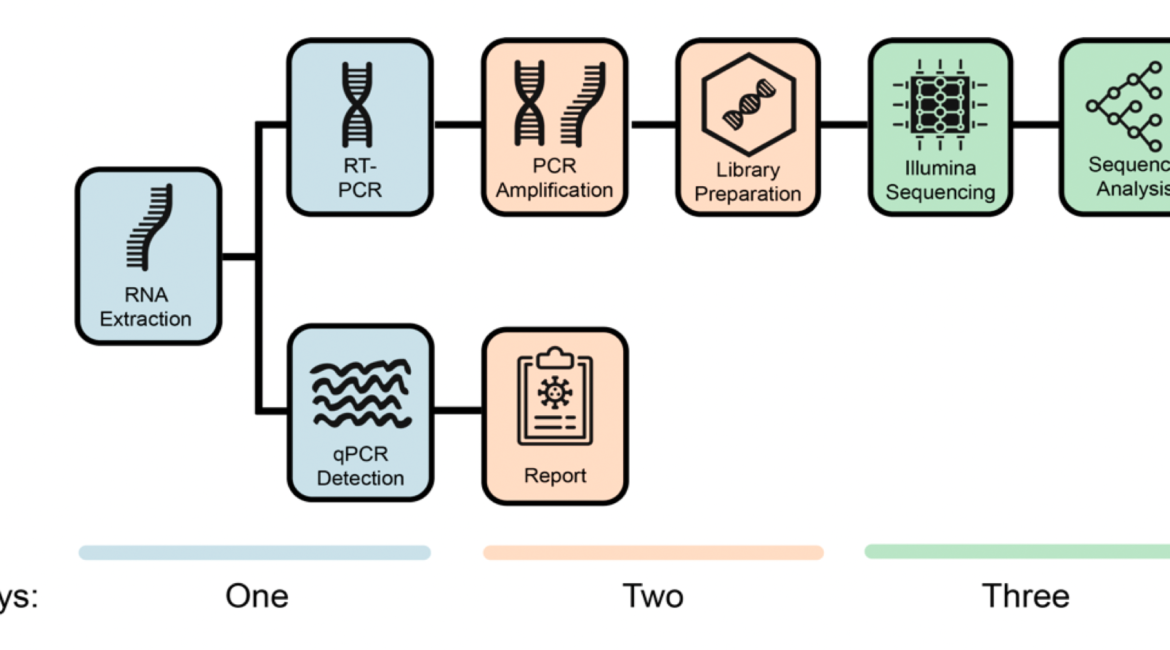
The COVID-19 pandemic spread very fast around the world. A few days after the first detected case in South Africa, an infection started a large hospital outbreak in Durban, KwaZulu-Natal. Phylogenetic analysis of SARS-CoV-2 genomes can be used to trace the path of transmission within a hospital. It can also identify the source of the outbreak and provide lessons to improve infection prevention and control strategies. In this manuscript, we outline the obstacles we encountered in order to genotype SARS-CoV-2 in real-time during an urgent outbreak investigation. In this process, we encountered problems with the length of the original genotyping protocol, reagent stockout and sample degradation and storage. However, we managed to set up three different library preparation methods for sequencing in Illumina. We also managed to decrease the hands on library preparation time from twelve to three hours, which allowed us to complete the outbreak investigation in just a few weeks. We also fine-tuned a simple bioinformatics workflow for the assembly of high-quality genomes in real-time. In order to allow other laboratories to learn from our experience, we released all of the library preparation and bioinformatics protocols publicly and distributed them to other laboratories of the South African Network for Genomics Surveillance (SANGS) consortium.