AIDS Res Hum Retroviruses, 2020.
Authors: Junqueira DM, Wilkinson E, Vallari AS, Deng XD, Achari A, Yu G, McArthur C, Kaptué L, Mbanya DN, Chiu C, Cloherty G, de Oliveira T, Rodgers MA.
Journal: AIDS Res Hum Retroviruses,doi: 10.1089/AID.2020.0031: (2020)
Abstract
Although the first HIV circulating recombinant form (CRF01_AE) is the predominant strain in many Asian countries, it is uncommonly found in the Congo Basin from where it first originated. In order to fill the gap in the evolutionary history of this important strain, we sequenced near complete genomes from HIV samples with subgenomic CRF01_AE regions collected in Cameroon and the Democratic Republic of the Congo (DRC) from 2001-2006.
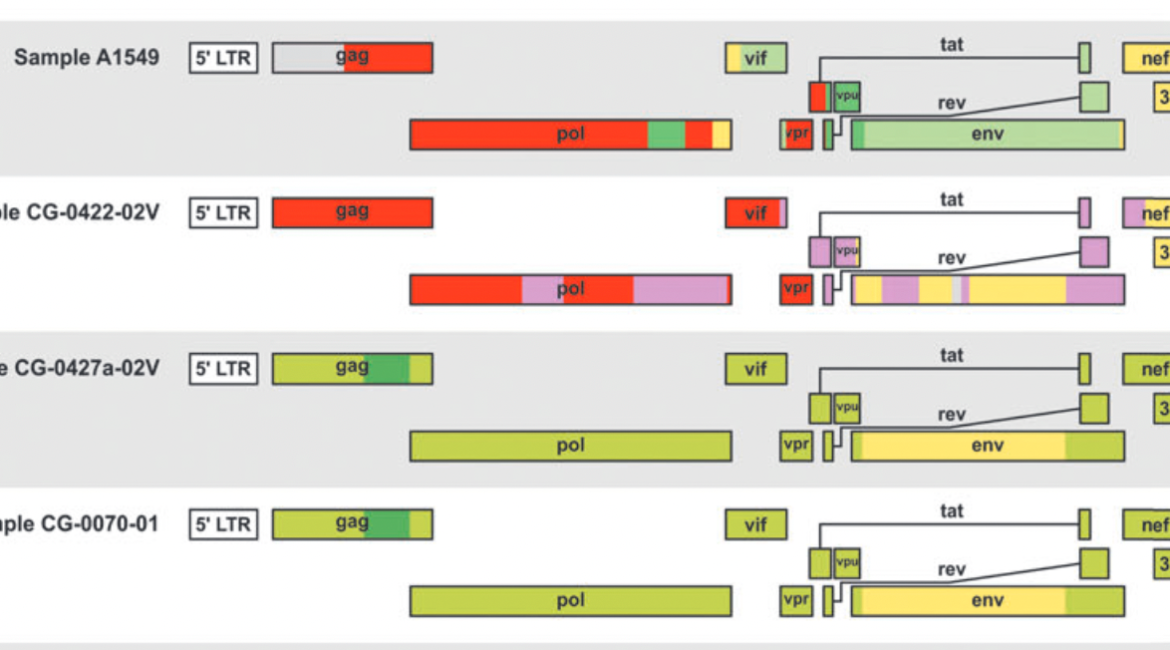
HIV genomes were generated from N=13 plasma specimens by next generation sequencing of metagenomic libraries prepared with spiked primers targeting HIV, followed by Sanger gap-filling. Genome sequences were aligned to reference strains, including Asian and African CRF01_AE sequences, and evaluated by phylogenetic and recombinant analysis to identify 4 CRF01_AE strains from Cameroon. We also identified 2 CRF02, 1 CRF27, and 6 unique recombinant form (URFs) genomes (01|A1|G, 01|02|F|U, F|G|01, A1|D|01, F|G|01, A1|G|01).
Phylogenetic analysis including the 4 new African CRF01_AE genomes placed these samples as a bridge between basal Central African Republic CRF01_AE strains and all Asian, European, and American CRF01_AE strains. Molecular dating confirmed previous estimates indicating that the most recent common CRF01_AE ancestor emerged in the early 1970s (1968 – 1970) and spread beyond Africa around 1980 to Asia. The new sequences and analysis presented here expands the molecular history of the CRF01_AE clade, and is illustrated in an interactive Next Strain phylogenetic tree, map, and timeline at (https://nextstrain.org/community/EduanWilkinson/hiv-1_crf01).