PLoS Negl Trop Dis, 2019.
Authors: Fonseca V, Libin PJK, Theys K, Faria NR, Nunes MRT, Restovic MI, Freire M, Giovanetti M, Cuypers L, Nowa A, Abecasis A, Deforche K, Santiago GA, Siqueira IC, San EJ, Machado KCB, Azevedo V, Filippis AMB, Cunha RVD, Pybus OG, Vandamme AM, Alcantara LCJ, de Oliveira T.
Journal: PLoS Negl Trop Dis,13(5):e0007231: doi: 10.1371/journal.pntd.0007231 (2019)
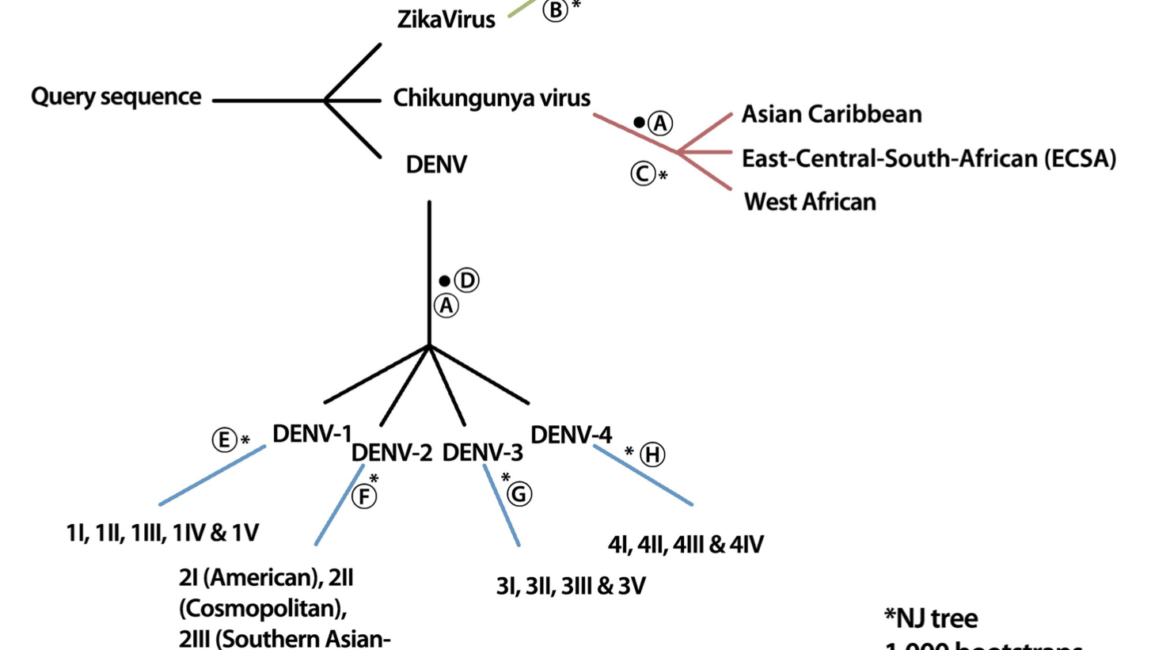
In recent years, an increasing number of outbreaks of Dengue, Chikungunya and Zika viruses have been reported in Asia and the Americas. Monitoring virus genotype diversity is crucial to understand the emergence and spread of outbreaks, both aspects that are vital to develop effective prevention and treatment strategies. Hence, we developed an efficient method to classify virus sequences with respect to their species and sub-species (i.e. serotype and/or genotype). This ArboTyping tool provides an easy-to-use software implementation of this new method and was validated on a large dataset assessing the classification performance with respect to whole-genome sequences and partial-genome sequences. Available online: http://www.krisp.org.za/tools.php