Lancet Microbe, 2020.
Authors: Msomi N, Mlisana K, Willianson C, Bhiman JN, Goedhals D, Engelbrecht S, Van Zyl G, Preiser W, Hardie D, Hsiao M, Mulder N, Martin D, Christoffels A, York D, Giandhari J, Wilkinson E, Pillay S, Tegally H, James SE, Kanzi A, Lessells RJ, de Oliveira T.
Journal: Lancet Microbe,https://doi.org/10.1016/S2666-5247(20)30116-6: (2020)
Abstract
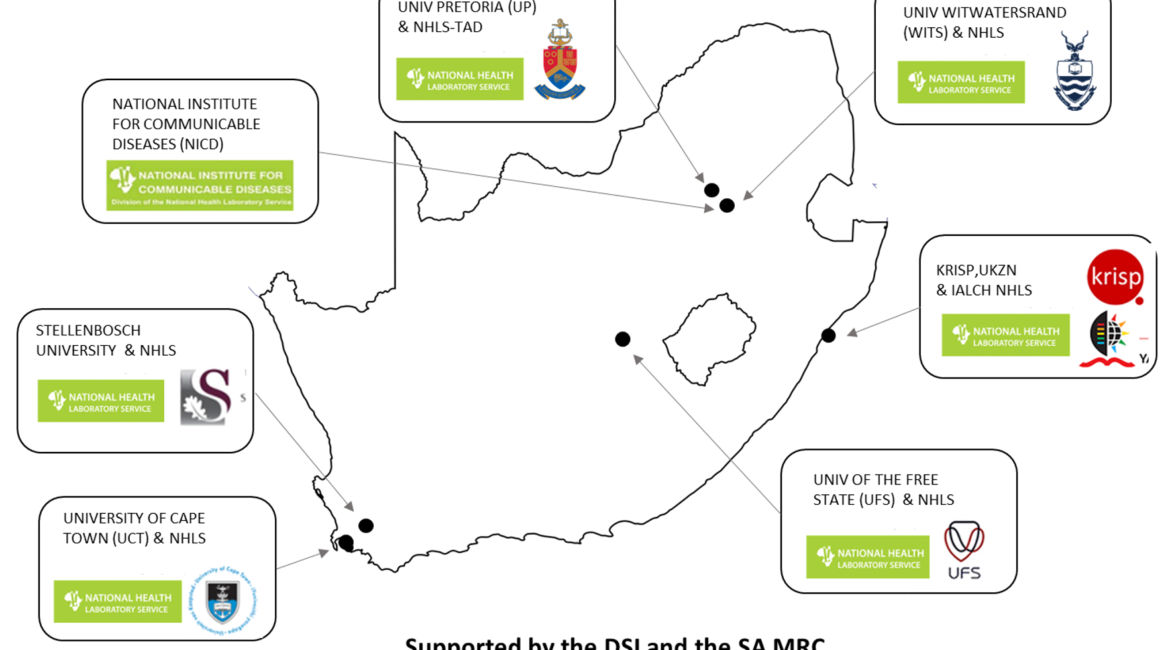
In response to the COVID-19 pandemic, we created the Network for Genomic Surveillance in South Africa (NGS-SA) in May, 2020 with grants from the South African Medical Research Council and the South African Department of Science and Innovation. Our goal is to sequence the genome of at least 10000 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) samples to inform the public health response in South Africa. As of July 27, 2020, we have sequenced the genome of 500 isolates, which has provided insights into how and when it was introduced into the country, and information on its early spread through clusters of outbreaks in health-care facilities, workplaces, and other congregate settings.
To respond to the virus, we have taken advantage of the public health research communities that exist in South Africa, and have established a laboratory network that, similar to the COVID-19 Genomics UK Consortium1 launched in March, 2020, will be guided by the following six key principles: (1) sequencing close to sample collection; (2) being platform agnostic; (3) supporting locally relevant public health priorities, such as by controlling clusters of outbreaks; (4) ensuring rapid and responsible open data sharing; (5) creating a bioinformatics system to process and analyse data locally; and (6) producing timely reports to inform policy makers. In addition, we have embedded a training and capacity-building programme for local scientists and health-care workers.